Fungal secondary metabolism: regulation, function and drug discovery
One of the exciting movements in microbial sciences has been a refocusing and revitalization of efforts to mine the fungal secondary metabolome. The magnitude of biosynthetic gene clusters (BGCs) in a single filamentous fungal genome combined with the historic number of sequenced genomes suggests that the secondary metabolite wealth of filamentous fungi is largely untapped. Mining algorithms and scalable expression platforms have greatly expanded access to the chemical repertoire of fungal-derived secondary metabolites. In this Review, I discuss new insights into the transcriptional and epigenetic regulation of BGCs and the ecological roles of fungal secondary metabolites in warfare, defence and development. I also explore avenues for the identification of new fungal metabolites and the challenges in harvesting fungal-derived secondary metabolites.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
206,07 € per year
only 17,17 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
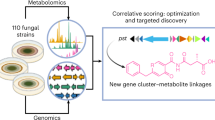
Correlative metabologenomics of 110 fungi reveals metabolite–gene cluster pairs
Article 06 March 2023
Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi
Article 26 April 2022
Engineered and total biosynthesis of fungal specialized metabolites
Article 03 January 2024
References
- Nesbitt, B. F., O’Kelly, J., Sargeant, K. & Sheridan, A. Aspergillus flavus and turkey X disease: toxic metabolites of Aspergillus flavus. Nature195, 1062–1063 (1962). CASPubMedGoogle Scholar
- Quinn, R. Rethinking antibiotic research and development: World War II and the penicillin collaborative. Am. J. Public Health103, 426–434 (2013). PubMedPubMed CentralGoogle Scholar
- Krause, D. J. et al. Functional and evolutionary characterization of a secondary metabolite gene cluster in budding yeasts. Proc. Natl Acad. Sci. USA115, 11030–11035 (2018). This study characterizes the pulcherrimin cluster inK. lactis, a yeast that belongs to a taxon not associated with secondary metabolism.CASPubMedGoogle Scholar
- Trail, F. et al. Physical and transcriptional map of an aflatoxin gene cluster in Aspergillus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Appl. Environ. Microbiol.61, 2665–2673 (1995). CASPubMedPubMed CentralGoogle Scholar
- Lind, A. L., Lim, F. Y., Soukup, A. A., Keller, N. P. & Rokas, A. An LaeA- and BrlA-dependent cellular network governs tissue-specific secondary metabolism in the human pathogen Aspergillus fumigatus. mSphere3, e00050–18 (2018). PubMedPubMed CentralGoogle Scholar
- Lysøe, E., Seong, K.-Y. & Kistler, H. C. The transcriptome of Fusarium graminearum during the infection of wheat. Mol. Plant Microbe Interact.24, 995–1000 (2011). PubMedGoogle Scholar
- Spraker, J. E. et al. Conserved responses in a war of small molecules between fungi and a bacterium. mBio9, e00820–18 (2018). The paper reports the conserved induction of an antibacterial secondary metabolite cluster across disparate fungal genera in response to a lipopeptide that is secreted by the invading bacterium. CASPubMedPubMed CentralGoogle Scholar
- Pelaez, F. in Handbook of Industrial Mycology (ed. Zhiqiang, A.) (Marcel Dekker, New York, NY, 2005).
- Schueffler, A. & Anke, T. Fungal natural products in research and development. Nat. Prod. Rep.31, 1425–1448 (2014). CASPubMedGoogle Scholar
- Kück, U., Bloemendal, S. & Teichert, I. Putting fungi to work: harvesting a cornucopia of drugs, toxins, and antibiotics. PLOS Pathog.10, e1003950 (2014). PubMedPubMed CentralGoogle Scholar
- Caldwell, G. A., Naider, F. & Becker, J. M. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev.59, 406–422 (1995). CASPubMedPubMed CentralGoogle Scholar
- Clevenger, K. D. et al. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat. Chem. Biol.13, 895–901 (2017). This paper presents a method to capture the entire secondary metabolome of a single species using FAC-MS technology. CASPubMedPubMed CentralGoogle Scholar
- Yun, C.-S., Motoyama, T. & Osada, H. Biosynthesis of the mycotoxin tenuazonic acid by a fungal NRPS-PKS hybrid enzyme. Nat. Commun.6, 8758 (2015). CASPubMedPubMed CentralGoogle Scholar
- Hur, G. H., Vickery, C. R. & Burkart, M. D. Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat. Prod. Rep.29, 1074 (2012). CASPubMedPubMed CentralGoogle Scholar
- Schmidt-Dannert, C. Biosynthesis of terpenoid natural products in fungi. Adv. Biochem. Eng. Biotechnol.148, 19–61 (2014). Google Scholar
- Li, X.-W., Ear, A. & Nay, B. Hirsutellones and beyond: figuring out the biological and synthetic logics toward chemical complexity in fungal PKS-NRPS compounds. Nat. Prod. Rep.30, 765 (2013). PubMedGoogle Scholar
- Chiang, Y.-M., Oakley, B. R., Keller, N. P. & Wang, C. C. C. Unraveling polyketide synthesis in members of the genus Aspergillus. Appl. Microbiol. Biotechnol.86, 1719–1736 (2010). CASPubMedPubMed CentralGoogle Scholar
- Umemura, M. et al. Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genet. Biol.68, 23–30 (2014). This study identifies the first BGC that produces a ribosomally encoded cyclic peptide. CASPubMedGoogle Scholar
- Pettit, R. K. Small-molecule elicitation of microbial secondary metabolites. Microb. Biotechnol.4, 471–478 (2011). CASPubMedPubMed CentralGoogle Scholar
- Lim, F. Y. et al. Fungal isocyanide synthases and xanthocillin biosynthesis in Aspergillus fumigatus. mBio9, e00785–18 (2018). This study identifies novel BGCs that contain isocyanide synthases. CASPubMedPubMed CentralGoogle Scholar
- Yu, J. et al. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol.70, 1253–1262 (2004). CASPubMedPubMed CentralGoogle Scholar
- Amaike, S., Affeldt, K. J. & Keller, N. P. in The Mycota: Agricultural Applications 2nd edn Vol. 11 (ed. Kempken, F.) 59–74 (Springer, Berlin, 2013).
- Neubauer, L., Dopstadt, J., Humpf, H.-U. & Tudzynski, P. Identification and characterization of the ergochrome gene cluster in the plant pathogenic fungus Claviceps purpurea. Fungal Biol. Biotechnol.3, 2 (2016). PubMedPubMed CentralGoogle Scholar
- Lebar, M. D. et al. Identification and functional analysis of the aspergillic acid gene cluster in Aspergillus flavus. Fungal Genet. Biol.116, 14–23 (2018). CASPubMedGoogle Scholar
- Keller, N. P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol.11, 671–677 (2015). CASPubMedPubMed CentralGoogle Scholar
- Wiemann, P. et al. Prototype of an intertwined secondary-metabolite supercluster. Proc. Natl Acad. Sci. USA110, 17065–17070 (2013). This report describes a supercluster in which the genes encoding the secondary metabolites fumagillin and pseurotin are intertwined. CASPubMedGoogle Scholar
- Andersen, M. R. et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl Acad. Sci. USA110, E99–E107 (2013). This study identifies non-contiguous members within a BGC through expression data. CASPubMedGoogle Scholar
- Ohsato, S. et al. Transgenic rice plants expressing trichothecene 3-O-acetyltransferase show resistance to the Fusarium phytotoxin deoxynivalenol. Plant Cell Rep.26, 531–538 (2007). CASPubMedGoogle Scholar
- Bradshaw, R. E. et al. Fragmentation of an aflatoxin-like gene cluster in a forest pathogen. New Phytol.198, 525–535 (2013). This study reports the fragmentation of a gene cluster dedicated to the production of a secondary metabolite. CASPubMedGoogle Scholar
- Lim, F. Y. & Keller, N. P. Spatial and temporal control of fungal natural product synthesis. Nat. Prod. Rep.31, 1277–1286 (2014). CASPubMedPubMed CentralGoogle Scholar
- Kalinina, S. A., Jagels, A., Cramer, B., Geisen, R. & Humpf, H.-U. Influence of environmental factors on the production of penitrems A–F by Penicillium crustosum. Toxins9, 210 (2017). PubMed CentralGoogle Scholar
- Hewage, R. T., Aree, T., Mahidol, C., Ruchirawat, S. & Kittakoop, P. One strain-many compounds (OSMAC) method for production of polyketides, azaphilones, and an isochromanone using the endophytic fungus Dothideomycete sp. Phytochemistry108, 87–94 (2014). CASPubMedGoogle Scholar
- Joffe, A. Z. & Lisker, N. Effects of light, temperature, and pH value on aflatoxin production in vitro. Appl. Microbiol.18, 517–518 (1969). CASPubMedPubMed CentralGoogle Scholar
- Lind, A. L., Smith, T. D., Saterlee, T., Calvo, A. M. & Rokas, A. Regulation of secondary metabolism by the Velvet complex is temperature-responsive in Aspergillus. G36, 4023–4033 (2016). CASPubMedGoogle Scholar
- Hagiwara, D. et al. Temperature during conidiation affects stress tolerance, pigmentation, and trypacidin accumulation in the conidia of the airborne pathogen Aspergillus fumigatus. PLOS ONE12, e0177050 (2017). PubMedPubMed CentralGoogle Scholar
- Berthier, E. et al. Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLOS Pathog.9, e1003289 (2013). CASPubMedPubMed CentralGoogle Scholar
- Nazari, L., Manstretta, V. & Rossi, V. A non-linear model for temperature-dependent sporulation and T-2 and HT-2 production of Fusarium langsethiae and Fusarium sporotrichioides. Fungal Biol.120, 562–571 (2016). CASPubMedGoogle Scholar
- Bazafkan, H. et al. SUB1 has photoreceptor dependent and independent functions in sexual development and secondary metabolism in Trichoderma reesei. Mol. Microbiol.106, 742–759 (2017). CASPubMedGoogle Scholar
- Bayram, O. et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science320, 1504–1506 (2008). This paper describes the identification of a conserved transcriptional complex that coordinates global regulation of secondary metabolism. CASPubMedGoogle Scholar
- Pruss, S. et al. Role of the Alternaria alternata blue-light receptor LreA (white-collar 1) in spore formation and secondary metabolism. Appl. Environ. Microbiol.80, 2582–2591 (2014). PubMedPubMed CentralGoogle Scholar
- Monroy, A. A., Stappler, E., Schuster, A., Sulyok, M. & Schmoll, M. A. CRE1-regulated cluster is responsible for light dependent production of dihydrotrichotetronin in Trichoderma reesei. PLOS ONE12, e0182530 (2017). PubMedPubMed CentralGoogle Scholar
- Purschwitz, J. et al. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol.18, 255–259 (2008). CASPubMedGoogle Scholar
- Calvo, A. M. & Cary, J. W. Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol.6, 62 (2015). PubMedPubMed CentralGoogle Scholar
- Kenne, G. et al. Activation of aflatoxin biosynthesis alleviates total ROS in Aspergillus parasiticus. Toxins10, 57 (2018). PubMed CentralGoogle Scholar
- Montibus, M., Pinson-Gadais, L., Richard-Forget, F., Barreau, C. & Ponts, N. Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol.41, 295–308 (2015). CASPubMedGoogle Scholar
- Fountain, J. C. et al. Oxidative stress and carbon metabolism influence Aspergillus flavus transcriptome composition and secondary metabolite production. Sci. Rep.6, 38747 (2016). CASPubMedPubMed CentralGoogle Scholar
- Macheleidt, J. et al. Regulation and role of fungal secondary metabolites. Annu. Rev. Genet.50, 371–392 (2016). CASPubMedGoogle Scholar
- Fernandes, M., Keller, N. P. & Adams, T. H. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol.28, 1355–1365 (1998). CASPubMedGoogle Scholar
- Brown, D. W. et al. Identification of a 12-gene fusaric acid biosynthetic gene cluster in Fusarium species through comparative and functional genomics. Mol. Plant Microbe Interact.28, 319–332 (2015). CASPubMedGoogle Scholar
- Yin, W.-B. et al. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus. Aspergillus fumigatus. J. Am. Chem. Soc.135, 2064–2067 (2013). CASPubMedGoogle Scholar
- Wiemann, P. et al. Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Aspergillus fumigatus. Front. Microbiol.5, 530 (2014). PubMedPubMed CentralGoogle Scholar
- Bergmann, S. et al. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl. Environ. Microbiol.76, 8143–8149 (2010). CASPubMedPubMed CentralGoogle Scholar
- Bok, J. W. & Keller, N. P. in TheMycota: Biochemistry and Molecular Biology 3rd edn Vol. 3 (ed. Hoffmeister, D.) 21–29 (Springer International, Switzerland, 2016).
- Chettri, P. & Bradshaw, R. E. LaeA negatively regulates dothistromin production in the pine needle pathogen Dothistroma septosporum. Fungal Genet. Biol.97, 24–32 (2016). CASPubMedGoogle Scholar
- Oakley, C. E. et al. Discovery of McrA, a master regulator of Aspergillus secondary metabolism. Mol. Microbiol.103, 347–365 (2017). CASPubMedGoogle Scholar
- Lim, F. Y., Ames, B., Walsh, C. T. & Keller, N. P. Co-ordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus. Cell. Microbiol.16, 1267–1283 (2014). CASPubMedGoogle Scholar
- Mulinti, P. et al. Accumulation of ergot alkaloids during conidiophore development in Aspergillus fumigatus. Curr. Microbiol.68, 1–5 (2014). CASPubMedGoogle Scholar
- Cichewicz, R. H. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep.27, 11–22 (2010). CASPubMedGoogle Scholar
- Roze, L. V., Arthur, A. E., Hong, S.-Y., Chanda, A. & Linz, J. E. The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol. Microbiol.66, 713–726 (2007). CASPubMedGoogle Scholar
- Shwab, E. K. et al. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell6, 1656–1664 (2007). CASPubMedPubMed CentralGoogle Scholar
- Gacek, A. & Strauss, J. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol.95, 1389–1404 (2012). CASPubMedPubMed CentralGoogle Scholar
- Fan, A. et al. Deletion of a histone acetyltransferase leads to the pleiotropic activation of natural products in Metarhizium robertsii. Org. Lett.19, 1686–1689 (2017). CASPubMedGoogle Scholar
- Gacek-Matthews, A. et al. KdmB, a Jumonji histone H3 demethylase, regulates genome-wide H3K4 trimethylation and is required for normal induction of secondary metabolism in Aspergillus nidulans. PLOS Genet.12, e1006222 (2016). Using genome-wide chromatin immunoprecipitation coupled with RNA-seq and liquid chromatography with tandem mass spectrometry (LC-MS/MS), this study presents unprecedented insight into the global epigenetic regulation of cryptic BGCs in one species. PubMedPubMed CentralGoogle Scholar
- Williams, R. B., Henrikson, J. C., Hoover, A. R., Lee, A. E. & Cichewicz, R. H. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem.6, 1895 (2008). CASPubMedGoogle Scholar
- Albright, J. C. et al. Large-scale metabolomics reveals a complex response of Aspergillus nidulans to epigenetic perturbation. ACS Chem. Biol.10, 1535–1541 (2015). CASPubMedPubMed CentralGoogle Scholar
- Reyes-Dominguez, Y. et al. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol.76, 1376–1386 (2010). CASPubMedPubMed CentralGoogle Scholar
- Karimi-Aghcheh, R. et al. Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G33, 369–378 (2013).
- Niehaus, E.-M. et al. Analysis of the global regulator Lae1 uncovers a connection between Lae1 and the histone acetyltransferase HAT1 in Fusarium fujikuroi. Appl. Microbiol. Biotechnol.102, 279–295 (2018). CASPubMedGoogle Scholar
- Nützmann, H.-W. et al. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc. Natl Acad. Sci. USA108, 14282–14287 (2011). This paper reports the bacterial induction of a cryptic BGC via a chromatin remodelling enzyme complex. PubMedGoogle Scholar
- Netzker, T. et al. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol.6, 299 (2015). PubMedPubMed CentralGoogle Scholar
- Bok, J. W. et al. VeA and MvlA repression of the cryptic orsellinic acid gene cluster in Aspergillus nidulans involves histone 3 acetylation. Mol. Microbiol.89, 963–974 (2013). CASPubMedGoogle Scholar
- Tsai, H. F., Wheeler, M. H., Chang, Y. C. & Kwon-Chung, K. J. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol.181, 6469–6477 (1999). This article presents the first identification of a BGC required for fungal development. CASPubMedGoogle Scholar
- Zhang, P. et al. A cryptic pigment biosynthetic pathway uncovered by heterologous expression is essential for conidial development in Pestalotiopsis fici. Mol. Microbiol.105, 469–483 (2017). CASPubMedGoogle Scholar
- Leonard, K. J. Virulence, temperature optima, and competitive abilities of isolines of races T and 0 of Bipolaris maydis. Phytopathology67, 1273–1279 (1977). Google Scholar
- Shukla, S. et al. Total phenolic content, antioxidant, tyrosinase and α-glucosidase inhibitory activities of water soluble extracts of noble starter culture Doenjang, a Korean fermented soybean sauce variety. Food Control59, 854–861 (2016). CASGoogle Scholar
- Eisenman, H. C. & Casadevall, A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol.93, 931–940 (2012). CASPubMedGoogle Scholar
- Jacobson, E. S. Pathogenic roles for fungal melanins. Clin. Microbiol. Rev.13, 708–717 (2000). CASPubMedPubMed CentralGoogle Scholar
- Zhao, L., Kim, J.-C., Paik, M.-J., Lee, W. & Hur, J.-S. A. Multifunctional and possible skin UV protectant, (3R)-5-hydroxymellein, produced by an endolichenic fungus isolated from Parmotrema austrosinense. Molecules22, 26 (2016). PubMed CentralGoogle Scholar
- Zheng, H. et al. Redox metabolites signal polymicrobial biofilm development via the NapA oxidative stress cascade in Aspergillus. Curr. Biol.25, 29–37 (2015). CASPubMedGoogle Scholar
- Scherlach, K. & Hertweck, C. Mediators of mutualistic microbe-microbe interactions. Nat. Prod. Rep.35, 303–308 (2018). CASPubMedGoogle Scholar
- Zeilinger, S. et al. Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol. Rev.40, 182–207 (2016). CASPubMedGoogle Scholar
- Rohlfs, M. Fungal secondary metabolite dynamics in fungus-grazer interactions: novel insights and unanswered questions. Front. Microbiol.5, 788 (2014). PubMedGoogle Scholar
- Partida-Martinez, L. P. & Hertweck, C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature437, 884–888 (2005). CASPubMedGoogle Scholar
- Scherlach, K., Busch, B., Lackner, G., Paszkowski, U. & Hertweck, C. Symbiotic cooperation in the biosynthesis of a phytotoxin. Angew. Chem. Int. Ed.51, 9615–9618 (2012). CASGoogle Scholar
- Spraker, J. E., Sanchez, L. M., Lowe, T. M., Dorrestein, P. C. & Keller, N. P. Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J.10, 2317–2330 (2016). CASPubMedPubMed CentralGoogle Scholar
- Khalid, S. et al. NRPS-derived isoquinolines and lipopetides mediate antagonism between plant pathogenic fungi and bacteria. ACS Chem. Biol.13, 171–179 (2018). CASPubMedGoogle Scholar
- Schumacher, J. et al. A functional bikaverin biosynthesis gene cluster in rare strains of Botrytis cinerea is positively controlled by VELVET. PLOS ONE8, e53729 (2013). CASPubMedPubMed CentralGoogle Scholar
- Campbell, M. A., Rokas, A. & Slot, J. C. Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biol. Evol.4, 289–293 (2012). PubMedPubMed CentralGoogle Scholar
- Oh, D.-C., Poulsen, M., Currie, C. R. & Clardy, J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol.5, 391–393 (2009). CASPubMedPubMed CentralGoogle Scholar
- Dhodary, B., Schilg, M., Wirth, R. & Spiteller, D. Secondary metabolites from Escovopsis weberi and their role in attacking the garden fungus of leaf-cutting ants. Chemistry24, 4445–4452 (2018). CASPubMedGoogle Scholar
- Tauber, J. P., Gallegos-Monterrosa, R., Kovács, Á. T., Shelest, E. & Hoffmeister, D. Dissimilar pigment regulation in Serpula lacrymans and Paxillus involutus during inter-kingdom interactions. Microbiology164, 65–77 (2018). CASPubMedGoogle Scholar
- Tauber, J. P., Schroeckh, V., Shelest, E., Brakhage, A. A. & Hoffmeister, D. Bacteria induce pigment formation in the basidiomycete Serpula lacrymans. Environ. Microbiol.18, 5218–5227 (2016). CASPubMedGoogle Scholar
- Fan, Y. et al. Regulatory cascade and biological activity of Beauveria bassiana oosporein that limits bacterial growth after host death. Proc. Natl Acad. Sci. USA114, E1578–E1586 (2017). This paper reports the finding that a fungus-derived antibacterial compound poisons the food supply to limit microbial competition. CASPubMedGoogle Scholar
- Drott, M. T., Lazzaro, B. P., Brown, D. L., Carbone, I. & Milgroom, M. G. Balancing selection for aflatoxin in Aspergillus flavus is maintained through interference competition with, and fungivory by insects. Proc. Biol. Sci.284, 20172408 (2017). This article provides evidence that a toxic secondary metabolite provides a fitness advantage to the fungus during confrontations with insects. PubMedPubMed CentralGoogle Scholar
- Dolan, S. K., O’Keeffe, G., Jones, G. W. & Doyle, S. Resistance is not futile: gliotoxin biosynthesis, functionality and utility. Trends Microbiol.23, 419–428 (2015). CASPubMedGoogle Scholar
- Teijeira, F. et al. The transporter CefM involved in translocation of biosynthetic intermediates is essential for cephalosporin production. Biochem. J.418, 113–124 (2009). CASPubMedGoogle Scholar
- Scharf, D. H. et al. Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J. Am. Chem. Soc.132, 10136–10141 (2010). CASPubMedGoogle Scholar
- Abe, Y. et al. Effect of increased dosage of the ML-236B (compactin) biosynthetic gene cluster on ML-236B production in Penicillium citrinum. Mol. Genet. Genomics268, 130–137 (2002). CASPubMedGoogle Scholar
- Yeh, H.-H. et al. Resistance gene-guided genome mining: serial promoter exchanges in Aspergillus nidulans reveal the biosynthetic pathway for fellutamide B, a proteasome inhibitor. ACS Chem. Biol.11, 2275–2284 (2016). This paper provides the first evidence that duplicated, resistant target genes within a BGC provide self-protection. CASPubMedGoogle Scholar
- Yue, Q. et al. Genomics-driven discovery of a novel self-resistance mechanism in the echinocandin-producing fungus Pezicula radicicola. Environ. Microbiol.20, 3154–3167 (2018). CASPubMedGoogle Scholar
- Yan, Y. et al. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature559, 415–418 (2018). This study describes a genomics approach to identify duplicated resistance genes and the discovery of a bioactive natural-product herbicide. CASPubMedPubMed CentralGoogle Scholar
- Hansen, B. G. et al. A new class of IMP dehydrogenase with a role in self-resistance of mycophenolic acid producing fungi. BMC Microbiol.11, 202 (2011). This study reports on the initial demonstration that a duplicated target gene within a BGC can provide resistance to the BGC product using a heterologous host. CASPubMedPubMed CentralGoogle Scholar
- Larkin, E. L., Dharmaiah, S. & Ghannoum, M. A. Biofilms and beyond: expanding echinocandin utility. J. Antimicrob. Chemother.73, i73–i81 (2018). CASPubMedGoogle Scholar
- Studt, L., Wiemann, P., Kleigrewe, K., Humpf, H.-U. & Tudzynski, B. Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl. Environ. Microbiol.78, 4468–4480 (2012). CASPubMedPubMed CentralGoogle Scholar
- Zhao, Y. et al. Production of a fungal furocoumarin by a polyketide synthase gene cluster confers the chemo-resistance of Neurospora crassa to the predation by fungivorous arthropods. Environ. Microbiol.19, 3920–3929 (2017). CASPubMedGoogle Scholar
- Schindler, D. & Nowrousian, M. The polyketide synthase gene pks4 is essential for sexual development and regulates fruiting body morphology in Sordaria macrospora. Fungal Genet. Biol.68, 48–59 (2014). CASPubMedGoogle Scholar
- Becker, J., Liermann, J. C., Opatz, T., Anke, H. & Thines, E. GKK1032A2, a secondary metabolite from Penicillium sp. IBWF-029-96, inhibits conidial germination in the rice blast fungus Magnaporthe oryzae. J. Antibiot.65, 99–102 (2012). CASPubMedGoogle Scholar
- Nielsen, J. C. et al. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat. Microbiol.2, 17044 (2017). CASPubMedGoogle Scholar
- Medema, M. H. et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol.11, 625–631 (2015). This article presents a community effort to standardize annotations and metadata on BGCs and their products. CASPubMedPubMed CentralGoogle Scholar
- Alberti, F., Foster, G. D. & Bailey, A. M. Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biotechnol.101, 493–500 (2017). CASPubMedGoogle Scholar
- Chavali, A. K. & Rhee, S. Y. Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Brief. Bioinform.19, 1022–1034 (2017). PubMed CentralGoogle Scholar
- Khaldi, N. et al. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol.47, 736–741 (2010). CASPubMedPubMed CentralGoogle Scholar
- Medema, M. H. et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res.39, W339–W346 (2011). CASPubMedPubMed CentralGoogle Scholar
- Galagan, J. E. et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature438, 1105–1115 (2005). CASPubMedGoogle Scholar
- Machida, M. et al. Genome sequencing and analysis of Aspergillus oryzae. Nature438, 1157–1161 (2005). PubMedGoogle Scholar
- Nierman, W. C. et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature438, 1151–1156 (2005). CASPubMedGoogle Scholar
- Mohimani, H. et al. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun.9, 4035 (2018). PubMedPubMed CentralGoogle Scholar
- Janevska, S. et al. Establishment of the inducible Tet-On system for the activation of the silent trichosetin gene cluster in Fusarium fujikuroi. Toxins9, 126 (2017). PubMed CentralGoogle Scholar
- Jiang, T. et al. Overexpression of the global regulator LaeA in Chaetomium globosum leads to the biosynthesis of chaetoglobosin Z. J. Nat. Prod.79, 2487–2494 (2016). CASPubMedGoogle Scholar
- Palonen, E. K. et al. Transcriptomic complexity of Aspergillus terreus Velvet gene family under the influence of butyrolactone I. Microorganisms5, 12 (2017). PubMed CentralGoogle Scholar
- Adnani, N., Rajski, S. R. & Bugni, T. S. Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod. Rep.34, 784–814 (2017). CASPubMedPubMed CentralGoogle Scholar
- Billingsley, J. M., DeNicola, A. B. & Tang, Y. Technology development for natural product biosynthesis in Saccharomyces cerevisiae. Curr. Opin. Biotechnol.42, 74–83 (2016). CASPubMedPubMed CentralGoogle Scholar
- He, Y. et al. Recent advances in reconstructing microbial secondary metabolites biosynthesis in Aspergillus spp. Biotechnol. Adv.36, 739–783 (2018). CASPubMedGoogle Scholar
- Yin, W.-B. et al. Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synth. Biol.2, 629–634 (2013). CASPubMedPubMed CentralGoogle Scholar
- Harvey, C. J. B. et al. HEx: a heterologous expression platform for the discovery of fungal natural products. Sci. Adv.4, eaar5459 (2018). This paper describes the tool and protocol development that led to the expression of 41 BGCs and 22 compounds in a yeast heterologous expression system. PubMedPubMed CentralGoogle Scholar
- Stepien, Ł. The use of Fusarium secondary metabolite biosynthetic genes in chemotypic and phylogenetic studies. Crit. Rev. Microbiol.40, 176–185 (2014). CASPubMedGoogle Scholar
- Khaldi, N., Collemare, J., Lebrun, M.-H. & Wolfe, K. H. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol.9, R18 (2008). This early phylogenetic study provides evidence for horizontal transfer of natural-product BGCs in fungi. PubMedPubMed CentralGoogle Scholar
- Reynolds, H. T. et al. Differential retention of gene functions in a secondary metabolite cluster. Mol. Biol. Evol.34, 2002–2015 (2017). CASPubMedGoogle Scholar
- Bignell, E., Cairns, T. C., Throckmorton, K., Nierman, W. C. & Keller, N. P. Secondary metabolite arsenal of an opportunistic pathogenic fungus. Phil. Trans. R. Soc. B371, 20160023 (2016). PubMedGoogle Scholar
- Perrin, R. M. et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLOS Pathog.3, e50 (2007). PubMedPubMed CentralGoogle Scholar
- Lind, A. L. et al. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLOS Biol.15, e2003583 (2017). This study compares DNA sequences of the BGCs of 66A. fumigatusisolates and establishes 5 drivers of genetic diversity that explain BGC macroevolutionary patterns.PubMedPubMed CentralGoogle Scholar
- Droce, A. et al. Functional analysis of the fusarielin biosynthetic gene cluster. Molecules21, 1710 (2016). PubMed CentralGoogle Scholar
- Campbell, M. A., Staats, M., van Kan, J. A. L., Rokas, A. & Slot, J. C. Repeated loss of an anciently horizontally transferred gene cluster in Botrytis. Mycologia105, 1126–1134 (2013). PubMedGoogle Scholar
- Nielsen, K. F. & Larsen, T. O. The importance of mass spectrometric dereplication in fungal secondary metabolite analysis. Front. Microbiol.6, 71 (2015). PubMedPubMed CentralGoogle Scholar
- Chiang, Y.-M. et al. Development of genetic dereplication strains in Aspergillus nidulans results in the discovery of aspercryptin. Angew. Chem. Int. Ed.55, 1662–1665 (2016). CASGoogle Scholar
- Díez, B. et al. The cluster of penicillin biosynthetic genes: identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J. Biol. Chem.265, 16358–16365 (1990). PubMedGoogle Scholar
- Smith, D. J., Burnham, M. K., Edwards, J., Earl, A. J. & Turner, G. Cloning and heterologous expression of the penicillin biosynthetic gene cluster from Penicillum chrysogenum. Biotechnology8, 39–41 (1990). CASPubMedGoogle Scholar
- Keller, N. P. & Hohn, T. M. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol.21, 17–29 (1997). CASPubMedGoogle Scholar
- Goffeau, A. et al. Life with 6000 genes. Science274, 546–567 (1996). CASPubMedGoogle Scholar
- Inglis, D. O. et al. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol.13, 91 (2013). CASPubMedPubMed CentralGoogle Scholar
- Samson, R. A. et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol.78, 141–173 (2014). CASPubMedPubMed CentralGoogle Scholar
- Visagie, C. M. et al. Identification and nomenclature of the genus Penicillium. Stud. Mycol.78, 343–371 (2014). CASPubMedPubMed CentralGoogle Scholar
- Kirk, P. M., Cannon, P. F., David, J. C. & Stalpers, J. A. (eds) Ainsworth & Bisby’s Dictionary of the Fungi 9th edn (CABI, 2001).
- Schoch, C. L. et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol.64, 1–15 (2009). CASPubMedPubMed CentralGoogle Scholar
- Jahn, L. et al. Linking secondary metabolites to biosynthesis genes in the fungal endophyte Cyanodermella asteris: the anti-cancer bisanthraquinone skyrin. J. Biotechnol.257, 233–239 (2017). CASPubMedGoogle Scholar
- Yin, W.-B. et al. An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol. Microbiol.83, 1024–1034 (2012). CASPubMedPubMed CentralGoogle Scholar
- Soukup, A. A. et al. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol. Microbiol.86, 314–330 (2012). CASPubMedPubMed CentralGoogle Scholar
- Itoh, E. et al. Sirtuin A regulates secondary metabolite production by Aspergillus nidulans. J. Gen. Appl. Microbiol.63, 228–235 (2017). CASPubMedGoogle Scholar
- Ahuja, M. et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J. Am. Chem. Soc.134, 8212–8221 (2012). CASPubMedPubMed CentralGoogle Scholar
- Brakhage, A. A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol.11, 21–32 (2013). CASPubMedGoogle Scholar
- Pfannenstiel, B. T. et al. Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. mBio8, e01246–17 (2017). CASPubMedPubMed CentralGoogle Scholar
- Gacek-Matthews, A. et al. KdmA, a histone H3 demethylase with bipartite function, differentially regulates primary and secondary metabolism in Aspergillus nidulans. Mol. Microbiol.96, 839–860 (2015). CASGoogle Scholar
- Strauss, J. & Reyes-Dominguez, Y. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol.48, 62–69 (2011). CASPubMedGoogle Scholar
- Wiemann, P. et al. CoIN: co-inducible nitrate expression system for secondary metabolites in Aspergillus nidulans. Fungal Biol. Biotechnol. 5, 6 (2018).
- Blin, K. et al. antiSMASH 4.0 — improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res.45, W36–W41 (2017). CASPubMedPubMed CentralGoogle Scholar
- Skinnider, M. A. et al. Genomes to natural products prediction informatics for secondary metabolomes (PRISM). Nucleic Acids Res.43, 9645–9662 (2015). CASPubMedPubMed CentralGoogle Scholar
- Wolf, T., Shelest, V., Nath, N. & Shelest, E. CASSIS and SMIPS: promoter-based prediction of secondary metabolite gene clusters in eukaryotic genomes. Bioinformatics32, 1138–1143 (2016). CASPubMedGoogle Scholar
- Vesth, T. C., Brandl, J. & Andersen, M. R. FunGeneClusterS: predicting fungal gene clusters from genome and transcriptome data. Synth. Syst. Biotechnol.1, 122–129 (2016). PubMedPubMed CentralGoogle Scholar
- Zierep, P. F. et al. SeMPI: a genome-based secondary metabolite prediction and identification web server. Nucleic Acids Res.45, W64–W71 (2017). CASPubMedPubMed CentralGoogle Scholar
- Conway, K. R. & Boddy, C. N. ClusterMine360: a database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res.41, D402–D407 (2013). CASPubMedGoogle Scholar
- Hadjithomas, M. et al. IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. mBio6, e00932 (2015). CASPubMedPubMed CentralGoogle Scholar
- Medema, M. H. et al. Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLOS Comput. Biol.10, e1003822 (2014). PubMedPubMed CentralGoogle Scholar
- Dejong, C. A. et al. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat. Chem. Biol.12, 1007–1014 (2016). CASPubMedGoogle Scholar
- Röttig, M. et al. NRPSpredictor2 — a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res.39, W362–W367 (2011). PubMedPubMed CentralGoogle Scholar
Acknowledgements
The author thanks F. Y. Lim for generating the original figure 5 and J. Winans and C. D. Nwagwu for help with formatting the text. N.P.K. is funded by US National Institutes of Health (NIH) grants R01GM112739-01 and R01 AI065728-01.









